research
The goal of our team is to leverage expertise from the field of single cell genomics in order to sequence tiniest amounts of RNA in diverse acellular contexts.
Condensate sequencing
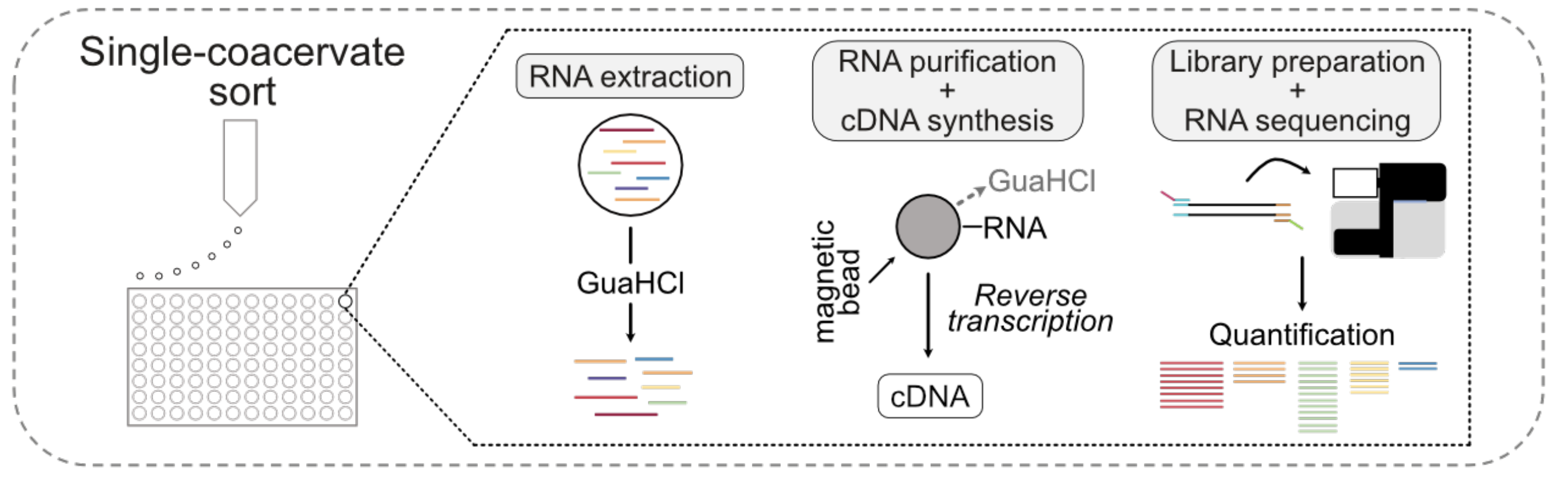
Liquid-liquid phase separation (LLPS) describes the demixing of polymers driven by macromolecular interactions. The resulting condensates have previously been used for encapsulation of small molecules, proteins, RNA, DNA, and other biomaterials for applications ranging from biomedical sensing to food production. More recently, phase separated microdroplets have been exploited as artificial cell mimics for biological systems due to their ease of self-assembly and their dynamic nature. In cells, LLPS organizes biomolecules into a wide range of membraneless organelles such as stress granules, P-bodies, Cajal bodies and many more. These membraneless compartments are involved in multiple cellular processes, ranging from regulation of gene expression over signaling cascades to stress adaptation. In order to comprehensively characterize the RNA content of phase separated condensates, we have previously developed an approach to read out RNA from single condensates. We adapted single-cell RNA sequencing technology in order to answer the question how different one condensate is from another in terms of RNA composition and how this relates to condensate features such as droplet size. We further determined that RNA molecules that enrich in condensates are characterized by specific sequence motifs strongly resembling short interspersed nuclear elements (SINEs). This approach therefore represents a powerful addition to labor intensive and low through microscopy approaches which previously have been the state-of-the-art approach for condensate RNA characterization. In the future, we aim to more deeply characterize the RNA content of different condensate types and in order to elucidate how condensate chemistry influences RNA assembly into condensates under different conditions.
Cell-free RNA sequencing
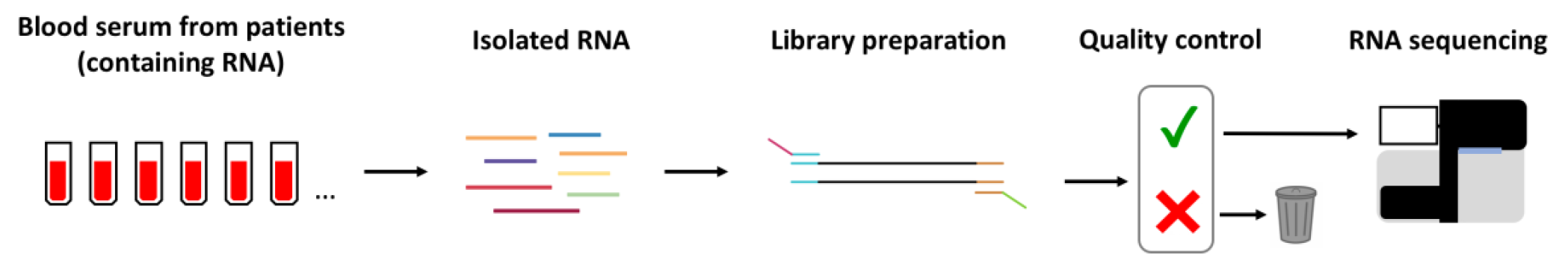
Cell-free RNA is present in the blood of every human being as a result of vesicular secretion from the cells of the human body. Sequencing cell-free RNA has been of very promising for the diagnosis of several diseases ranging from cancer to cardiovascular diseases. In contrast to protein-based biomarkers, a huge advantage of RNA is that it can be amplified. In recent years technological developments enabled the amplification and sequencing of tiny amounts of RNA even from single cells. Another advantage of RNA over DNA (which could also be amplified) is that it is continuously shed from cells. In contrast DNA exits cells only when the cell is dying. Hence, cell-free RNA holds the promise of continuous probing of transcriptional changes in the cells of the human body. We aim to apply methods from the field of single cell RNA sequencing to sequence cell-free RNA in different medically relevant contexts. This approach enables the sequencing of minute amounts of RNA and our expertise in sequencing very small amounts of RNA from cells or even phase separated condensates will facilitate the detection of even smallest amounts of RNA from the blood or other